ISLES 2016: Now online!
Important Notices
- The challenge article was accepted by Medical Image Analysis and can now be found online. Please refer to this work if you publish any result obtained on the ISLES 2015 data or want to refer to it.
- The final rankings can be found under results. We would like to thank all the participants for their hard work as well as the invited speakers for their interesting talks. The challenge has been a great success!
- We are proud to welcome a total of 20 participating teams
- Bern decided to change the SPES evaluation scheme to the “penumbra”-label only
- We are happy to announce that the highest ranking and particularly interesting methods will be invited to contribute to the workshops LNCS post-proceedings volume.
- Information: invited speakers, proceedings (v2015-09-30), on-site program (v2015-09-30)
- Links: evaluation code, MICCAI registration
- See VSD for registration, training data download, test data download and evaluation system.
Overview
Welcome to Ischemic Stroke Lesion Segmentation (ISLES), a medical image segmentation challenge at the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) 2015 (October 5-9th). We aim to provide a platform for a fair and direct comparison of methods for ischemic stroke lesion segmentation from multi-spectral MRI images. A public dataset of diverse ischemic stroke cases and a suitable automatic evaluation procedure will be made available for the two following tasks:
SISS: sub-acute ischemic stroke lesion segmentation
SPES: acute stroke outcome/penumbra estimation
On this page, you can find details on how the challenge works in the about section, the clinical and research motivation behind it, the rules of how to participate, the data description and the evaluation scheme.
Dates
All dates are subject to change, please keep an eye on the important notices.
| Official launch of the website | |
| Registration opens and distribution of training data | |
| Opening of the evaluation system | |
| Early-bird reduced registration fees | |
| Short paper submission deadline 11:59 am (GMT) |
|
| Distribution of test data | |
| Deadline for submitting test data results | |
| MICCAI 2015 workshop and award presentation | |
| Compilation of high-impact publication & LNCS post-proceedings invitation |
|
| Training data and evaluation are made public |
Results
The frozen ranking of the teams who participated at the ISLES workshop, held the 5th of October 2015 at the MICCAI 2015, is presented here and constitutes the final outcome of the challenges. Running results of the continued challenge can be found on the VSD data/evaluation web-page. Please note that the results there might vary from the ones presented in this table, as at the VSD page, the ASSD and HD are computed over all but empty segmentation masks, irregardless of the DC score.
SISS
| rank2 | first author (VSD-name) & affiliation | cases | ASSD1 | DC | HD1 | |
|---|---|---|---|---|---|---|
| 7.54 | Liang Chen (chenl2) Biomedical Image Analysis Group, Imperial College London |
34/36 | 11.71±10.12 | 0.44±0.30 | 70.61±24.59 |  |
| 7.66 | Francis Dutil (dutif1) Université de Sherbrooke, Sherbrooke |
27/36 | 9.25±9.79 | 0.35±0.32 | 44.91±32.53 |  |
| 3.82 | Chaolu Feng (fengc1) College of Inform. Science and Eng., Northeastern University, Shenyang |
32/36 | 3.27±3.62 | 0.55±0.30 | 19.78±15.65 |  |
| 9.21 | Michael Goetz (goetm2) Junior Group Med. Img. Comp., German Cancer Research Center (DKFZ), Heidelberg |
35/36 | 14.20±10.41 | 0.33±0.28 | 77.95±22.13 |  |
| 7.92 | Tom Haeck (haect1) ESAT/PSI, Dept. of Electrical Engineering, KU Leuven |
30/36 | 12.24±13.49 | 0.37±0.33 | 58.65±29.99 |  |
| 5.63 | Hanna Halme (halmh1) HUS Medical Imaging Center, University of Helsinki and Helsinki University Hospital |
31/36 | 8.05±9.57 | 0.47±0.32 | 40.23±33.17 |  |
| 7.97 | Andrew Jesson (jessa3) Centre for Intelligent Machines, McGill University |
31/36 | 11.04±13.68 | 0.32±0.26 | 40.42±26.98 |  |
| 3.25 | Konstantinos Kamnitsas (kamnk1) Biomedical Image Analysis Group, Imperial College London |
34/36 | 5.96±9.38 | 0.59±0.31 | 37.88±30.06 |  |
| 9.18 | Mahmood Qaiser (mahmq2) Signals and Systems, Chalmers University of Technology, Gothenburg |
30/36 | 10.00±6.61 | 0.38±0.28 | 72.16±17.32 |  |
| 6.70 | Oskar Maier (maieo1) Institute of Medical Informatics, Universität zu Lübeck |
31/36 | 10.21±9.44 | 0.42±0.33 | 49.17±29.6 |  |
| 7.07 | John Muschelli (muscj1) Johns Hopkins Bloomberg School of Public Health |
33/36 | 11.54±11.14 | 0.42±0.32 | 62.43±28.64 |  |
| 6.40 | Syed Reza (rezas1) Vision Lab, Old Dominion University, Norfolk |
33/36 | 6.24±5.21 | 0.43±0.27 | 41.76±25.11 |  |
| 6.67 | David Robben (robbd1) ESAT/PSI, Dept. of Electrical Engineering, KU Leuven |
33/36 | 11.27±10.17 | 0.43±0.30 | 60.79±31.14 |  |
| 10.99 | Ching-Wei Wang (wangc2) Grad. Inst. of Biomed. Eng., National Taiwan University of Science and Technology |
15/36 | 7.59±6.24 | 0.16±0.26 | 38.54±20.36 |  |
SPES
| rank2 | first author (VSD-name) & affiliation | cases | ASSD1 | DC | |
|---|---|---|---|---|---|
| 2.02 | Richard McKinley (mckir1) Dep. of Diag. and Interventional Neuroradiology, Inselspital, Bern Uni. Hospital |
20/20 | 1.65±1.40 | 0.82±0.08 |  |
| 2.20 | Oskar Maier (maieo1) Institute of Medical Informatics, Universität zu Lübeck |
20/20 | 1.36±0.74 | 0.81±0.09 |  |
| 3.92 | David Robben (robbd1) ESAT/PSI, Dept. of Electrical Engineering, KU Leuven |
20/20 | 2.77±3.27 | 0.78±0.09 |  |
| 4.05 | Chaolu Feng (fengc1) College of Inform. Science and Eng., Northeastern University, Shenyang |
20/20 | 2.29±1.76 | 0.76±0.09 |  |
| 4.60 | Elias Keller (kelle1) Department of Radiology, Medical Physics, University Medical Center Freiburg |
20/20 | 2.44±1.93 | 0.73±0.13 |  |
| 5.15 | Tom Haeck (haect1) ESAT/PSI, Dept. of Electrical Engineering, KU Leuven |
20/20 | 4.00±3.39 | 0.67±0.24 |  |
| 6.05 | Francis Dutil (dutif1) Université de Sherbrooke, Sherbrooke |
20/20 | 5.53±7.59 | 0.54±0.26 |  |
1Computed excluding the failed cases (where DC=0, see cases-column), therefore not suitable for a direct comparison of methods
2Relative rank, computed as described below in the evaluation section
Each row is linked to a method description as PDF file. The complete proceedings can be found here (v2015-09-30)
Photos
Conclusion
The BrainLes workshop took place at the 5th of October 2015. In front of a full room of roughly one hundred listeners, the first three of the invited speakers commenced with their lectures: Maria Rocca highlighted the latest developments in Multiple Sclerosis treatment, Arya Nabavi helped improving our understanding of stroke from a medical perspective and James Fishbaught (for Guido Gerig) talked about imaging of Trauma Brain Injuries. Then the first batch of contributed papers were presented.
After a short break, Simon Warfield entertained us with an overview over his long line of work on label fusion, followed by the BRATS challenge.
Afterwards, it was time for the ISLES challenge. First, Bianca de Haan promoted the neuropsychologic perspective on stroke lesion segmentation and detailed the requirements arising from the field, which differ from the medical point of view. Then we gave a short introduction of the challenges' tasks and announced the winners (Congratulations!). These then had the opportunity to present their methods in form of a short presentation. Finally, a first interpretation of the ISLES results was given.
In the afternoon, the workshop continued with a talk of Roland Wiest on Advanced Neuroimaging for Glimo. After a few more participating works, a lively discussion arose, sprinkled with much welcome recommendations for any future challenge.
We would like to thank the participants for their interest and hard work, the invited speakers for their valuable contributions and the audience for their numerous appearance and the lively discussions. It has been a great experience with all of you!
About
Why challenges?
Medical image processing comprises a large number of tasks, for which new methods are regularly proposed. But, more often than desirable, it is difficult to compare their fitness, as the reported results are obtained on private datasets. Challenges aim to overcome these shortcomings by providing (1) a public dataset that reflects the diversity of the problem and (2) a platform for a fair and direct comparison of methods with suitable evaluation measures. Thus, the scientific progress is promoted (see grand-challenge.org for a detailed rationale behind challenges).
Past challenges
During the recent years, the need for challenges has been emphasized in the medical image processing community. Especially in the scope of the MICCAI and International Symposium on Biomedical Imaging (ISBI) conferences, a number of challenges have been already successfully organized. All of these are collected centrally at grand-challenges.org.
The ISLES challenge
With ISLES, we provide such a challenge covering ischemic stroke lesion segmentation in multi-spectral MRI data. The task is backed by a well established clinical and research motivation and a large number of already existing methods. Each team may participate in either one or both of two sub-tasks: sub-acute ischemic stroke lesion segmentation (SISS) and acute stroke outcome/penumbra estimation (SPES).
SISS: Automatic segmentation of ischemic stroke lesion volumes from multi-spectral MRI sequences acquired in the sub-acute stroke development stage.
SPES: Automatic segmentation of acute ischemic stroke lesion volumes from multi-spectral MRI sequences for stroke outcome prediction.
How it works
Interested participants can download a set of training cases (data description) with associated expert segmentations of the stroke lesions to train and evaluate their approach. Shortly before MICCAI 2015 takes place, a set of test cases will be made available and the participants upload their segmentation results (in the form of binary images) as well as a short paper describing their method. The organizers then evaluate each case and establish a ranking of the participating teams. At the MICCAI 2015 conference, the results will be presented in the scope of a workshop and the results discussed together with invited experts. Each team will present their method as a poster and, furthermore, a number of selected teams will have the opportunity to hold a small presentation detailing their method. A number of high ranking and particularly interesting approaches will be invited to contribute to the workshops LNCS post-proceedings volume. After the challenge, the organizers and participants will together compose a high-impact journal paper to publish their findings.
Motivation
Stroke is the second most frequent cause of death and a major cause of disability in industrial countries. In patients who survive, stroke is often associated with high socioeconomic costs due to persistent disability. Its most frequent manifestation is the ischemic stroke, triggered by local thrombosis, hemodynamic factors or embolic causes, whose diagnosis often involves the acquisition of brain magnetic resonance (MR) scans to assess the stroke lesion presence, location, extent, evolution and other factors. An automated method to locate, segment and quantify the lesion area would support clinicians and researchers, rendering their findings more robust and reproducible. See Rekik et al. (2012) for a plea for automatic stroke lesion segmentation and Maier et al. (2014) [preprint] or Kabir et al. (2007) for recent methods. See also the comparison study of stroke segmentation methods conducted by the challenge authors: Maier et al. (2015) [preprint].
Clinical motivation
To assess stroke treatments, such as thrombolytic therapy or embolectomy, and to discover suitable markers for sound treatment decisions, large clinical studies tracking the stroke development and outcome under controlled conditions are necessary. Automatic stroke segmentation facilitates the quantification of the stroke lesion over time. The same holds true for quantitative studies aiming to answer the questions 'how an ischemic stroke evolves' and to discover the involved processes.
In cases of acute ischaemic stroke, advanced neuroimaging techniques are recommended for a quick, reliable diagnosis and stratification for therapy, which is typically done by assessing the tissue-at-risk. The infarct core (tissue which is already infarcted, or is irrevocably destined to become so) can be identified using diffusion-weighted magnetic resonance imaging. Meanwhile, surrounding tissue that can be potentially rescued (the “penumbra”) is characterized by hypo-perfusion in perfusion-weighted MRI. The imaging is used to assess the ratio of tissue that could potentially be saved, to tissue which is inevitably destined to infarct. Currently this is performed "by eye" by neuroradiologists, or using off the shelf tools that segment by simple thresholding.
Research motivation
Lesion to symptom mapping is a neurosciencentific approach used to detect correlations between brain areas and cognitive functions by means of negative samples. It requires manual segmentation of pathologies such as stroke lesions in many MR volumes, which is subject to high inter- and intra-observer variability, diminishing the validity of any gained insight as well as the reproducibility. Furthermore, an automatic method would enable the fully automatic screening of routinely acquired scans. The resulting lesion volume could then be mapped to functional and/or anatomical brain areas, for example to identify suitable study candidates.
Rules
What is required to participate?
Each team wishing to participate in the ISLES challenge is required to:
- Register at the data distribution and evaluation platform (see participate for more details).
- Upload the test data segmentation results before the deadline (see dates).
- Submit a short paper describing their method and their results on the training data set (see short paper).
- Register to the associated MICCAI 2015 workshop and pay the attendance fee (possible right up until the day of the challenge).
- Be present at the workshop with at least one team member and present a poster.
Which methods are called for?
All types of automatic stroke lesion segmentation methods are welcome. We specifically do not restrict the participation to new, innovative and unpublished methods, but invite research groups from all over the world to enter their existing method in the competition. Semi-automatic methods are eligible for participation and will appear in the ranking, but out of competition, as it is impossible to rate the influence of the manual steps in a fair manner.
Participating in one or both of the sub-task (SISS & SPES)
We encourage all teams to apply their method to both of the sub-tasks to highlight its generalization ability. But it is equally possible to submit a specialized approach for each of the task or just to one of them.
Organizational structure
ISLES is organized as part of a larger, three-part, full-day workshop at the MICCAI 2015 with the following organizational structure:
The three events should be regarded as a single workshop and a single registration allows to visit all of these and to participate in them. Please note that each part has its own rules of active participation, which can be found on their respective web-pages. The schedule can be found here, more information will follow.
Participation in the Brain Tumor Image Segmentation (BRATS) challenge
The organizers of ISLES and BRATS took care to apply the same pre-processing steps to their data, enabling the participants of one of the challenges to furthermore participate in the respectivly other. Especially machine learning based approaches should be easy to adapt. Double contributions are highly welcome and only one registration at the MICCAI 2015 workshop is required.
Usage of ISLES and BRATS data for the Brain Lesion Segmentation (BrainLeS) workshop
Participants are free to use the training data from both, ISLES and BRATS, to evaluate methods submitted to BrainLeS, but are then encouraged to also participate in the respective challenge. In such case, they are not required to submit the short-paper to ISLES respectively BRATS, instead their (accepted) BrainLeS manuscript will suffice, as long as it states the results obtained on the training set.
Short paper
ISLES requires the participating teams to submit a short-paper of 4 pages in LNCS format, which will be reviewed by the organizers. The text will be distributed among the MICCAI 2015 attendees on the MICCAI 2015 USB drive and uploaded to the challenge result web-page. See dates for the associated deadlines.
Requirements
- Compiled with LaTeX using the LNCS format (download the template)
- Clear, concise and detailed description of applied methods including all parameter values
- Motivation for choosing the described method is welcome
- Results on training set may be reported, but are not required
- The dataset should not be described
- Treat it more as a method description over four pages than as an actual article
Review
- We will only look for language and reproducibility
- Novelty or results do not matter
Submission
- Until the 10th of August 2015
- Via e-mail to maier@imi.uni-luebeck.de
- Hand in a PDF (with standard fonts) AND the complete sources.
- Stating "ISLES 2015" and you team name in the the subject
- Including the following copyright statement in the body: "I am the corresponding author of the paper
and in the name of all co-authors I declare that MICCAI has the right to distribute the submitted material to MICCAI members and workshop / challenge / tutorial and MICCAI attendees."
On-site method presentation
Poster
- each participating team presents its work as a poster
- preferred poster size is A0
- maximum poster size is 90cm x 120cm
- please include the ISLES logo to make your posters affiliation directly recognizable: blue, white, svg
Presentation
- four selected methods will be invited to give an oral presentation
- selected teams will be announced directly after the evaluation round
- 8+2 scheme: eight minutes presentation plus two minutes discussion
- a poster presentation is still mandatory
Post-proceedings publication (LNCS)
Some selected methods from the BRATS and ISLES challenge will be called upon to contribute an article to the BrainLes workshops Springer LNCS post-proceedings special issue with a longer version of their article. The respective participants are informed during the challenge and given a few weeks after the MICCAI to compose their article, which will have to undergo another review cycle.
Submitting results for multiple methods
A team can submit results from multiple methods, but an extra registration to the MICCAI 2015 challenge is required for each additionally submitted result.
Privacy and data copyright
Since the workshop, the data has been re-released under the Open Database License. Any rights in individual contents of the database are licensed under the Database Contents License.
In a nutshell: Please feel free to use the data in your research. We only would like to ask you to refer to the Challenge in your publication and to cite one of our works. In case you have the old version of the data under the challenge-only ISLES license, simply download the datasets again under the new license.
Data
The data for both sub-tasks, SISS and SPES, are pre-processed in a consistent manner to allow easy application of a method to both problems.
Image format
Uncompressed Neuroimaging Informatics Technology Initiative (NIfTI) format: *.nii.
(I/O libraries: MedPy (Python), NiftiLib (C,Java,Matlab,Python), Nibabel (Python), Tools for NIfTI and ANALYZE image (Matlab), ITK (C++))
Pre-processing
All MRI sequences are skull-stripped, re-sampled to an isotropic spacing of \(1^3 mm\) (SISS) resp. \(2^3 mm\) (SPES) and co-registered to the FLAIR (SISS) and T1w contrast (SPES) sequences respectively.
Data details
Data layout
SISS
Each case is distributed in its own folder (e.g. 01), containing the MRI sequences following the naming convention VSD.Brain.XX.O.MR_<sequence>.<VSDID>.000.nii. For the training cases, the folder will also contain a an expert segmentation image named VSD.Brain.XX.O.OT.<VSDID>.000.nii.
SPES
Each case is distributed in its own folder (e.g. Nr10), containing the MRI sequences following the naming convention VSD.Brain.XX.O.MR_<sequence>.<VSDID>.000.nii. For the training cases, the folder will also contain three sub-folders (corelabel, penumbralabel, mergedlabels) with the ground-truth labels, but note that only the "penumbra" label will be employed in the challenge.
Case characteristics
SISS
For SISS, cases with sub-acute ischemic stroke lesions have been collected from clinical routine acquisition. The following characteristics reflect the diversity of the included cases:
| Lesion count | \(\mu=2.49\) |
| \([1, 14]\) | |
| Lesion volume | \(\mu=17.59ml\) |
| \([0.001, 346.064]\) | |
| Haemorrhage present | \(n=12\) |
| 0=yes, 1=no | |
| Non-stroke white matter lesion load | \(\mu=1.34\) |
| 0=none, 1=small, 2=medium, 3=large | |
| Lesion localization (lobes) | \(n_1=11,n_2=24,n_3=42,n_4=17,n_5=2,n_6=6\) |
| 1=frontal, 2=temporal, 3=parietal, 4=occipital, 5=midbrain, 6=cerebellum | |
| Lesion localization (cortical/subcortical) | \(n_1=36,n_2=49\) |
| 1=cortical, 2=subcortical | |
| Affected artery | \(n_1=6, n_2=45, n_3=11, n_4=5, n_5=0\) |
| 1=ACA, 2=ACM, 3=ACP, 4=BA, 5=other | |
| Midline shift observable | \(n_0=51,n_1=5,n_2=0\) |
| 0=none, 1=slight, 2=strong | |
| Ventricular enhancement observable | \(n_0=38,n_1=15,n_2=3\) |
| 0=none, 1=slight, 2=strong | |
| Laterality | \(n_1=18,n_2=35,n_3=3\) |
| 1=left, 2=right, 3=both |
All presented statistics are for the 56 case from Lübeck spread over training and testing together. After the challenge more detailed statistics will be revealed.
SPES
Coming soon.
Participate
Data distribution, registration and automatic evaluation will be handled by the Virtual Skeleton Database:
There you will find explanations on how to register, how to download the data and how as well as in which format to upload your results. Furthermore, the evaluation scores obtained by each team will be listed there.
Evaluation (SISS & SPES)
The evaluation comprises three evaluation measures, each of them highlighting different aspects of the segmentation quality. From the computed evaluation scores, the global ranking of each team is established as described below.
Segmentation task
SISS
Automatic segmentation of the infarcted area in multi-spectral scans of sub-acute ischemic stroke cases. Success will be measured by the evaluation measures detailed below.
SPES
Automatic segmentation of the "penumbra" in multi-spectral scans of acute ischemic stroke cases. Success will be measured, as for SISS, by the evaluation measures detailed below. Training and testing data are released with an infarct core (diffusion restriction) and a merged label. These are not used in the challenge but can be employed for private evaluations, e.g. of perfusion-diffusion mismatches.
Expert segmentations
All expert segmentation were prepared by experienced raters. For SISS, two ground-truth sets will be made available, for SPES one. The segmentation guidelines for the expert raters were as follows:
SISS
Segmentation was conducted completely manual by an experienced MD. Lesions were classified as sub-acute infarct if pathologic signal was found concomitantly in Flair and DWI images (presence of vasogenic and cytotoxic edema with evidence of swelling due to increased water content). Infarct lesions with signal change due to hemorrhagic transformation were included. Acute infarct lesions (DWI signal for cytotoxic edema only, no FLAIR signal for vasogenic edema) or residual infarct lesions with gliosis and scarring after infarction (no DWI signal for cytotoxic edema, no evidence of swelling) were not included.
SPES
The label for the perfusion-restriction was segmented by a MD as follows: First, a semi-manual segmentation of the region of interest was done using the currently accepted TMax threshold of 6 seconds (Olivot et al, Stroke 2009; Straka et al., J Magn Reson Imaging 2010; Lansberg et al., Lancet Neurol 2012; Mishra et al., Stroke 2014; Wheeler et al., Stroke 2013) for the production of a preliminary label. Then, morphological operations were applied to the label in order to reduce interpolation artifacts and to correct for unphysiological outliers within the label. Afterwards, the label was hand-corrected again. As a final step, non-brain areas inside the label, such as sulci, ventricles and other CSF collections, were erased from the label. This label represents an underperfused area of potentially salvageable tissue at risk based on the TMax Map (calculated by PMA Assist, segmented in Slicer). TMax is the time it takes for the tissue residue function to reach it's maximum value. TMax is a sensitive parameter reflecting changes in the perfusion state of brain tissue.
The label for the diffusion-restriction was segmented by a MD as follows: A semi-manual segmentation of the region of interest was done using the currently accepted ADC threshold of \(600\times 10^-6 mm^2/sec\) (Straka et al., J Magn Reson Imaging 2010, Lansberg et al., Lancet Neurol 2012, Mishra et al., Stroke 2014, Wheeler et al., Stroke 2013). This label represents an area with impairment of energy metabolism and serves as a surrogate to identify the ischemic core of the lesion, which is deemed to undergo infarction.
The resulting penumbra (target mismatch = perfusion-restriction label minus diffusion-restriction label) is in line with the MRI profile that suggests salvageable tissue and a favorable response to re-perfusion treatment applied in the multi-center DEFUSE and DEFUSE-II trial (Albers et al., Ann Neurol 2006; Lansberg et al., Lancet Neurol 2012).
Note that only the "penumbra" label will be employed in the challenge.
How it works
Data upload
To upload your training or testing segmentations, navigate to https://www.virtualskeleton.ch/ISLES/Start2015 and select the respective evaluation button, which will lead you to a page where you can 1. Upload, 2. Evaluate and 3. Activate your segmentations. All of these steps are described in more detail on the evaluation page. Please take care to (1) comply with the required naming format and (2) give your segmentations expressive names to be able to distinguish between the versions. New and improved segmentations can be uploaded at any time, without any restrictions.
Training cases
All segmentations selected under "Finished segmentations" will appear in the "Competitive segmentations" listing and and a summary of the results is displayed on the main page (https://www.virtualskeleton.ch/ISLES/Start2015), which allows you to compare your results against the competitors. Please be aware that we do not provide a ranking for the training results, as they can easily be tuned and are of limited significance. Likewise, do not be intimidated by teams with very good results - you never now how they might have come to pass and how their method will perform on the testing set.
Shortly, a new functionality will allow you to download the detailed case-wise results of each team as CSV file to perform your own analyses offline.
Testing cases
Uploading the segmentation results for the testing set will work similar as for the training data, only you won't be able to see the evaluation results, nor will there be a summary posted on the main page. Thus, tuning of the results should be prevented. Each teams results and their ranking will be revealed in-place during the MICCAI 2015. Afterwards, the results will furthermore be placed on the challenge websites in a frozen table.
After the challenge
After the challenge, both evaluation systems will stay open. The only difference being that the testing cases will equally be evaluated and displayed immediately. Hence, later results are prone to tuning.
Evaluation measures
Three principal evaluation measures are employed to evaluate the quality of a segmentation compared to the reference ground-truth: The Dice's coefficient (DC) denotes the volume overlap, the average symmetric surface distance (ASSD) the surface fit and the Hausdorff distance (HD) the maximum error. Furthermore, precision & recall are provided to reveal over- respectively under-segmentation, but will not be used in the computation of the team ranks, as they are indirectly reflected by the DC value.
Dice's coefficient (DC)
The \(DC\) measures the similarity between two datasets. Considering two sets of volume voxels \(A\) and \(B\), the \(DC\) value is given as:
$$DC=\dfrac{2|A\cap B|}{|A|+|B|}$$
A value of 0 indicates no overlap, a value of 1 perfect similarity. During interpretation, it should be kept in mind that the \(DC\) is known to yield higher values for larger volumes i.e. a \(DC\) of 0.9 for lung segmentation is considered average, as is a \(DC\) of 0.6 for multiple sclerosis lesions.
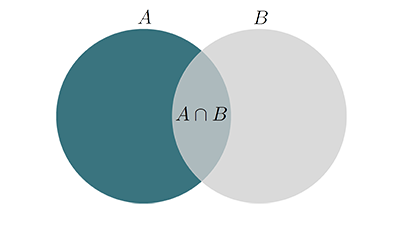 Fig.: The two sets \(A\), \(B\) and their union.
Fig.: The two sets \(A\), \(B\) and their union.
Note that the \(DC\) is the same as the F1-score, monotonic in Jaccard similarity and the harmonic mean of precision & recall. See here for more details.
Average symmetric surface distance (ASSD)
The \(ASSD\) denotes the average distance between the volumes surface points averaged over both directions. Considering two sets of surface points \(A\) and \(B\), the average surface distance (\(ASD\)) is given as:
$$ASD(A,B) = \frac{\sum_{a\in A}min_{b\in B}d(a,b)}{|A|}$$
with \(d(a,b)\) being the Euclidean distance between the points \(a\) and \(b\). Since \(ASD(A,B)\neq ASD(B,A)\), the \(ASSD\) is then given as:
$$ASSD(A,B) = \frac{ASD(A,B)+ASD(B,A)}{2}$$
It is given in \(mm\), the lower the better, and works equally well for large and small objects.
Hausdorff distance (HD)
The \(HD\) denotes the maximum distance between two volumes surface points and hence denotes outliers, especially when multiple objects are considered. It is defined as:
$$HD(A,B)=\text{max}\{\max_{a\in A}\min_{b\in B}d(a,b),\max_{b\in B}\min_{a\in A}d(b,a)\}$$
Similar to the \(ASSD\), the \(HD\) is given in \(mm\) and a lower value denotes a better segmentation.
Precision & Recall
The \(precision\) (also called positive predictive value) and \(recall\) (also known as sensitivity) of two sets are defined as
$$precision=\dfrac{TP}{TP+FP}$$ and $$recall=\dfrac{TP}{TP+FN}$$
where \(TP\) (true positive) denotes the overlapping points, \(FP\) (false positive) the wrong segmented points in the segmentation and \(FN\) (false negative) the not segmented points in the reference.
Both measure take values in the range of \([0,1]\). A relatively high precision compared to the recall reveals under-segmentation and vice-versa, as depicted in the following figure:
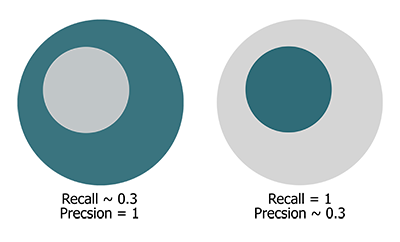 Fig.: Gray circle: Segmented points. Green circle: Reference points.
Fig.: Gray circle: Segmented points. Green circle: Reference points.
Ranking
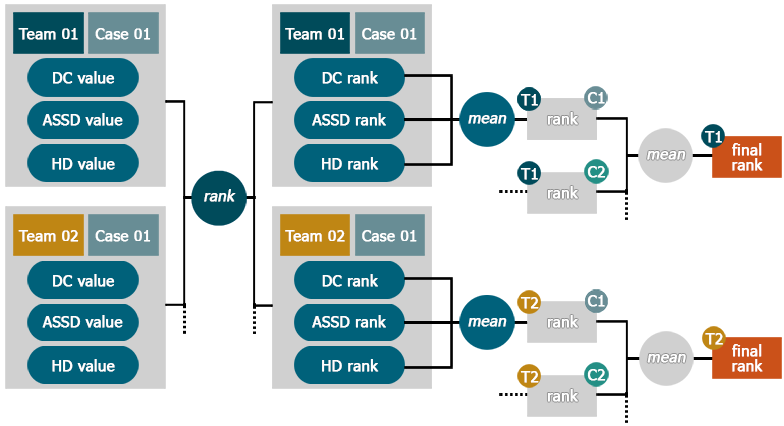
To establish a team's rank, the following steps are performed:
- Compute the \(DC\), \(ASSD\) & \(HD\) values for each case
- Establish each team's rank for \(DC\), \(ASSD\) & \(HD\) separately for each dataset
- Compute the mean rank over all three evaluation measures/case to obtain the team's rank for the case
- Compute the mean over all case-specific ranks to obtain the team's final rank
This scheme is furthermore detailed in the following figure:
Very easy or difficult cases
By not averaging the actual evaluation measures, but rather the ranks in each, the system is not affected by very difficult or easy cases: If all methods return low results on one case, even a small difference between them will still count in the ranking. Thus, the overall impression of a method will not be distorted.
Missing or not processable cases
If a case has not been uploaded or could not be processed for some reason, the associated team will be assigned the last place for this case.
Complete mismatch
A complete mismatch (i.e. a \(DC\) value of 0) will be seen as a missing case and treated as detailed above (the obtained \(ASSD\) & \(HD\) values are then ignored).
Shared ranks
Multiple teams can share a single rank. In such a cases, the following rank will be left empty.
SISS specifics
Since we provide two ground-truth segmentations for the testing data set, a rank is computed for both and then average over both of them to obtain the final rank.
SPES specifics
Since the ground-truth of SPES hold missing pixels, we decided to exclude the HD from the computation of the final ranking, as this measure would not fulfill its purpose of denoting the outliers.
Organizers
This challenge is organized by
Maier, Oskar, Institute of Medical Inform., Universität zu Lübeck (directed by Prof. Dr. rer. nat. habil. Heinz Handels)
Menze, Björn, Computer Science, Technische Universität München
Reyes, Mauricio, Institute for Surgical Technology & Biomechanics, Universität Bern.
Contributors
- Image data preparation
- Laszlo Friedmann, Institute of Medical Informatics, Universität zu Lübeck
- Web-page design
- Laszlo Friedmann, Institute of Medical Informatics, Universität zu Lübeck
- SISS image data source I
- Peter Schramm, Institute for Neuroradiology, Universitätsklinikum Schleswig-Holstein, Lübeck
Christian Erdmann, Institute for Neuroradiology, Universitätsklinikum Schleswig-Holstein, Lübeck - SISS image data source II
- Clinic for Neuroradiology, Technische Universität München
- SPES image data source
- Institute for Neuroradiology, Universitätsspital Bern
- Data download management
- Michael Kistler, The Virtual Skeleton Database
Roman Niklaus, The Virtual Skeleton Database - Evaluation management
- Michael Kistler, The Virtual Skeleton Database
Roman Niklaus, The Virtual Skeleton Database - Expertise and support
- Ulrike Krämer, Department of Neurology, Universitätsklinikum Schleswig-Holstein, Lübeck
André Kemmling, Institute for Neuroradiology, Universitätsklinikum Schleswig-Holstein, Lübeck - Expert segmentations SISS I
- Matthias Liebrand, Department of Neurology, Universitätsklinikum Schleswig-Holstein, Lübeck
- Expert segmentations SISS II
- Stefan Winzeck, Technische Universität München
- Expert segmentations SPES
- Levin Häni, Universitätsspital Bern
Supporters
- Graduate School for Computing in Medicine and Life Sciences, Universität zu Lübeck, Germany