Data
Image format
Uncompressed Neuroimaging Informatics Technology Initiative (NIfTI) format: *.nii.
(I/O libraries: MedPy (Python), NiftiLib (C,Java,Matlab,Python), Nibabel (Python), Tools for NIfTI and ANALYZE image (Matlab), ITK (C++))
Pre-processing
All MRI sequences are skull-stripped and anonymization was performed. No other pre-processing, such as re-sampling or co-registration is performed. This allows each participant to employ their favorite algorithms.
For the skull stripping of the structural images the stripTs tool was utilized. For the PWI images, a brain mask was created with the BET algorithm from FSL and the resulting mask was applied to the corresponding PWI image with the fslmath tool from FSL. Varying hand crafted parameters were chosen for the BET algorithm, depending on the respective MRI sequence. The decision of using two different algorithms for the skull stripping rests upon qualitative assessment.
To facilitate the participation, we decided to provide the data in two variants: A version with all sequences co-registered and skull-stripped with a common brain mask; and a version without co-registration and with individual skull-stripping performed. This enabled the participants to apply their favorite registration method, if desired.
Data details
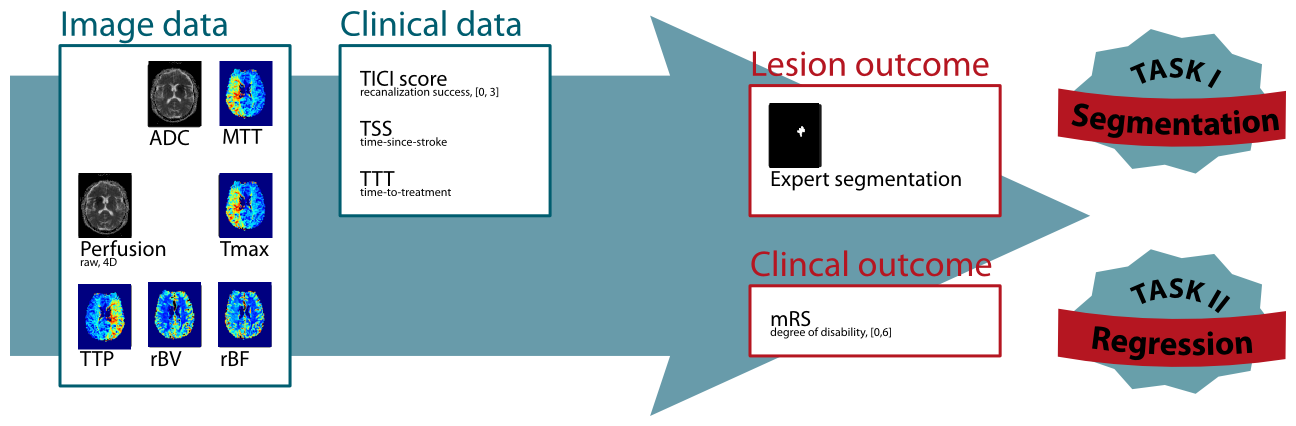
For ISLES 2016, 35 training and roughly 40 testing cases will be made public and can be downloaded from the SMIR platform after registration.
Clinical parameter details
Thrombolysis in Cerebral Infarction (TICI) scale
The TICI scale, originally proposed by Higashida et al. (2003), provides a standardized method to evaluate intracranial perfusion assessed in cerebral angiography. Here, it is used to assess the achieve re-perfusion after a flow-restoration intervention such as thrombectomy.
| Grade 0 |
No Perfusion. No antegrade flow beyond the point of occlusion. |
| Grade 1 |
Penetration With Minimal Perfusion. The contrast material passes beyond the area of obstruction but fails to opacify the entire
cerebral bed distal to the obstruction for the duration of the angiographic run. |
| Grade 2 |
Partial Perfusion. The contrast material passes beyond the obstruction and opacifies the arterial bed distal to the obstruction.
However, the rate of entry of contrast into the vessel distal to the obstruction and/or its rate of clearance from the distal bed are
perceptibly slower than its entry into and/or clearance from comparable areas not perfused by the previously occluded vessel, eg,
the opposite cerebral artery or the arterial bed proximal to the obstruction. |
| Grade 2a |
Only partial filling (2/3) of the entire vascular territory is visualized. |
| Grade 2b |
Complete filling of all of the expected vascular territory is visualized, but the filling is slower than normal. |
| Grade 3 |
Complete Perfusion. Antegrade flow into the bed distal to the obstruction occurs as promptly as into the obstruction
and clearance of contrast material from the involved bed is as rapid as from an uninvolved other bed of the same vessel or the opposite cerebral artery. |
Modified Rankin Scale (mRS)
The 90 days mRS is a standardized scale to assess the degree of disability 90 days after a stroke incidence and can be considered as the clinical outcome (van Swieten et al. (1988), see here for an online version of the test). The mRS score serves as endpoint for Task II.
| Grade 0 |
No Symptoms at all. |
| Grade 1 |
No significant disability despite symptoms; able to carry out all usual duties and activities. |
| Grade 2 |
Slight disability; unable to carry out all previous activities, but able to look after own affairs without assistance. |
| Grade 3 |
Moderate disability; requiring some help, but able to walk without assistance. |
| Grade 4 |
Moderately severe disability; unable to walk without assistance and unable to attend to own bodily needs without assistance. |
| Grade 5 |
Severe disability; bedridden, incontinent and requiring constant nursing care and attention. |
| Grade 6 |
Dead. |
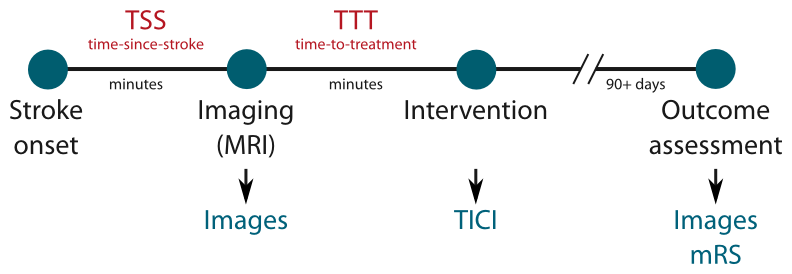
Time-since-stroke (TSS)
TSS is an important factor as only a fast re-establishing of the cerebral perfusion can lead to benefits for the patient. The less time passed since stroke onset, the more likely a re-perfusion can salvage brain tissue.
Time-to-treatment (TTT)
TTT denotes the time passed between imaging the patient and the re-perfusion treatment. Similar to the TSS, its value determines the amount of salvageable brain tissue.
Acute imaging details
For this challenge, MRI sequences recorded at the time of admission are as detailed below. For more information on the sequences, see e.g. http://mriquestions.com.
ADC map
Constructed from the DWI images, the ADC map show the underperfused area as hypointense region. Compared to the DWI trace images, the ADC map does not suffer from confounding T2 shine-through effects.
Raw perfusion data
Perfusion imaging (here: dynamic susceptibility imaging) denotes the perfusion of brain tissue with blood over time. The raw perfusion data are multiple 3D scans performed at equal time-intervals combined into a 4D volume.
Perfusion maps (MTT, Tmax, TTP, rBV, rBF)
For clinical assessment various maps can be computed from the raw perfusion data, each of which denotes different aspects of the measured brain perfusion. For ISLES 2016, a number of maps computed via block decomposition using the Olea Sphere software are provided.
Follow-up stroke imaging
To assess the final lesion outcome, an anatomical sequence is acquired when the stroke lesion stabilized. The manually segmented lesion in this scan constitutes the final lesion outcome and serves as end-point for Task I.